While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Biotechnology
Unlocking the Cell’s Energy Hub: Scientists Reveal How ATP Enters the Endoplasmic Reticulum
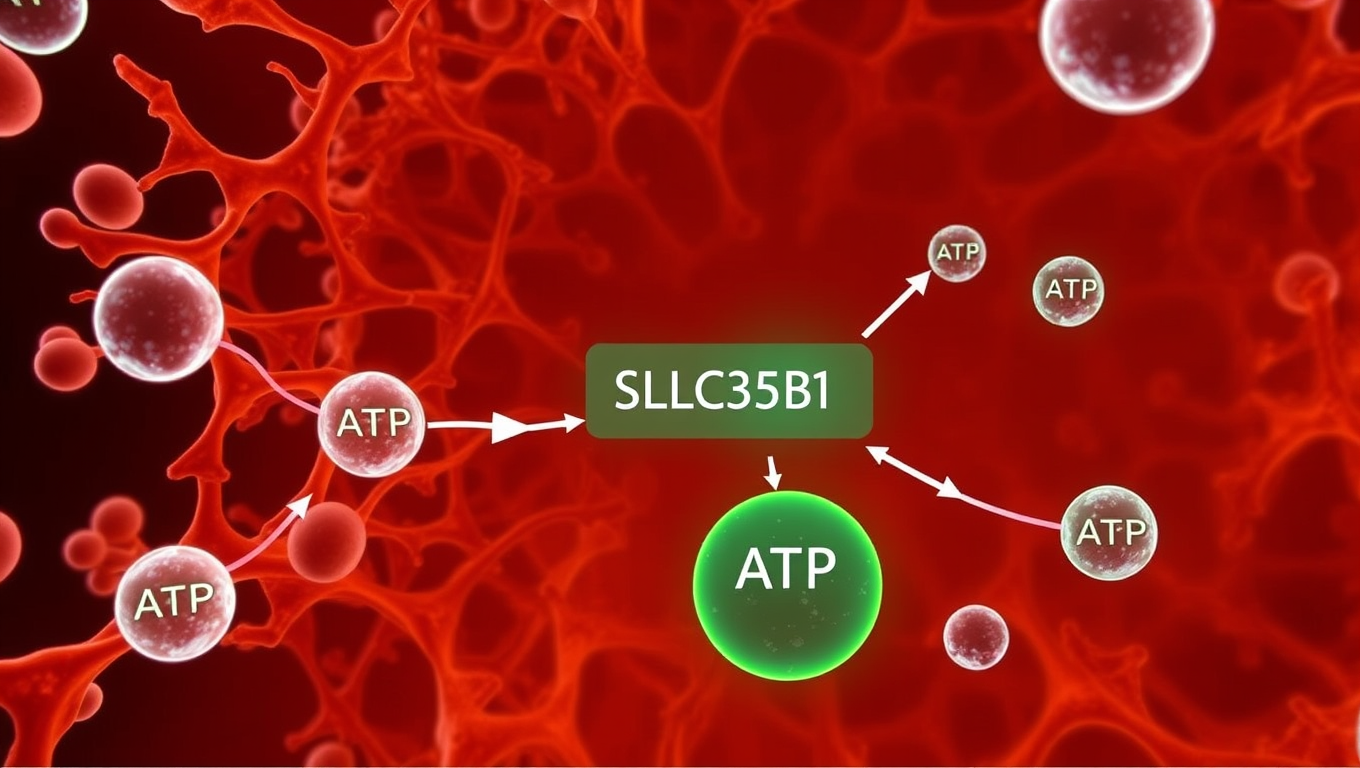
A team of scientists has answered a long-standing question in cell biology, uncovering how the cell’s main energy currency, ATP, is transported into the endoplasmic reticulum (ER). Disrupted energy transport could affect diseases such as type 2 diabetes, cancer, and neurodegenerative disorders. The study confirms that the transporter protein SLC35B1 is the key gateway for ATP entry into the ER.
Behavioral Science
“Rewired for Romance: Scientists Give Gift-Giving Behavior to Singing Fruit Flies”
By flipping a single genetic switch, researchers made one fruit fly species adopt the gift-giving courtship of another, showing how tiny brain rewiring can drive evolutionary change.
Agriculture and Food
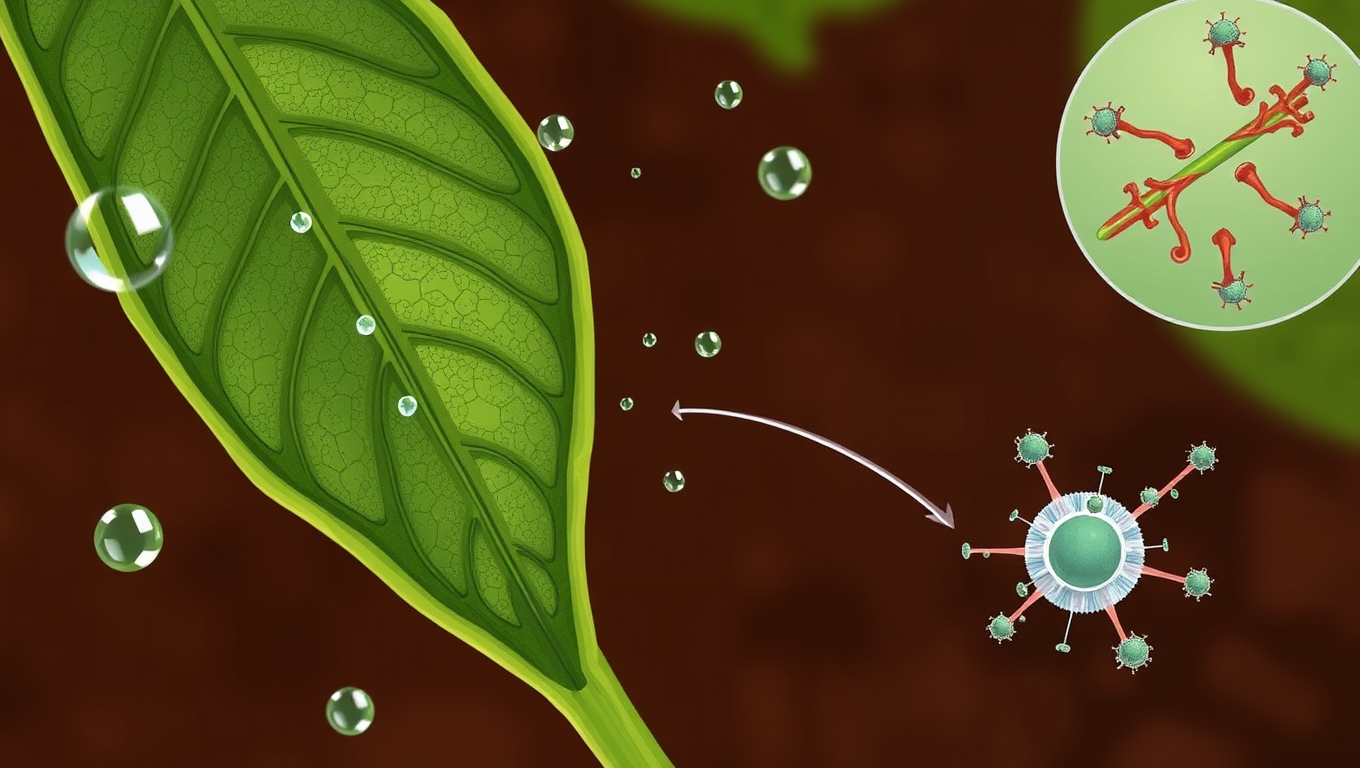
The Secret Motor Protein That Saves Crops from Drought: Uncovering Myosin XI’s Role in Plant Stress Response
Scientists have discovered that a protein once thought to be just a cellular “courier” actually helps plants survive drought. This motor protein, myosin XI, plays a critical role in helping leaves close their pores to conserve water. When it’s missing, plants lose water faster, respond poorly to drought, and activate fewer protective systems. The finding could open the door to hardier crops that can withstand a warming, drying world.
Biochemistry Research
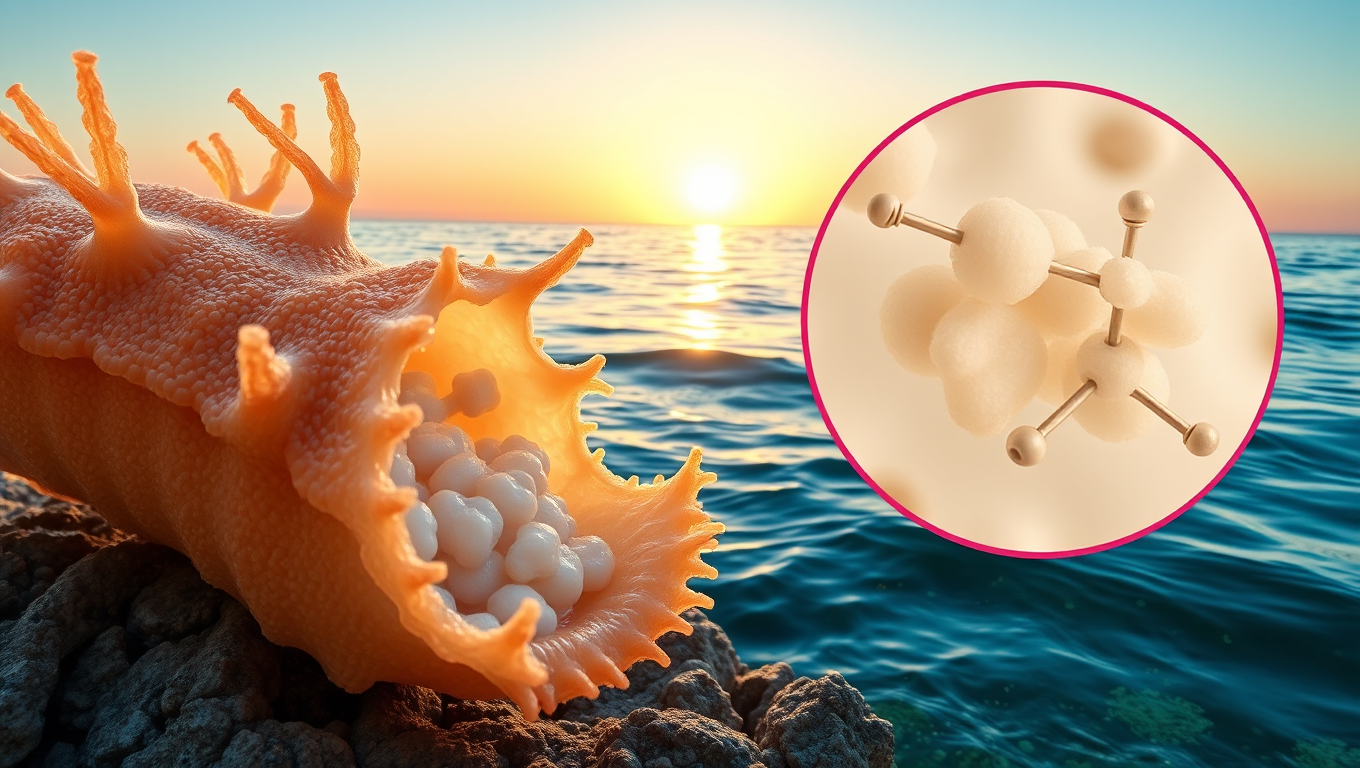
Unlocking Nature’s Secrets: Scientists Discover Natural Cancer-Fighting Sugar in Sea Cucumbers
Sea cucumbers, long known for cleaning the ocean floor, may also harbor a powerful cancer-fighting secret. Scientists discovered a unique sugar in these marine creatures that can block Sulf-2, an enzyme that cancer cells use to spread. Unlike traditional medications, this compound doesn t cause dangerous blood clotting issues and offers a cleaner, potentially more sustainable way to develop carbohydrate-based drugs if scientists can find a way to synthesize it in the lab.
-

 Detectors11 months ago
Detectors11 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate12 months ago
Earth & Climate12 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer12 months ago
Cancer12 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein12 months ago
Albert Einstein12 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry11 months ago
Chemistry11 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate12 months ago
Earth & Climate12 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food12 months ago
Agriculture and Food12 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions12 months ago
Diseases and Conditions12 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention