While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Bacteria
“Revealing Bacteria’s Hidden Past: How Machine Learning Uncovered a Timeline of Evolutionary Milestones”
Scientists have helped to construct a detailed timeline for bacterial evolution, suggesting some bacteria used oxygen long before evolving the ability to produce it through photosynthesis.

Allergy
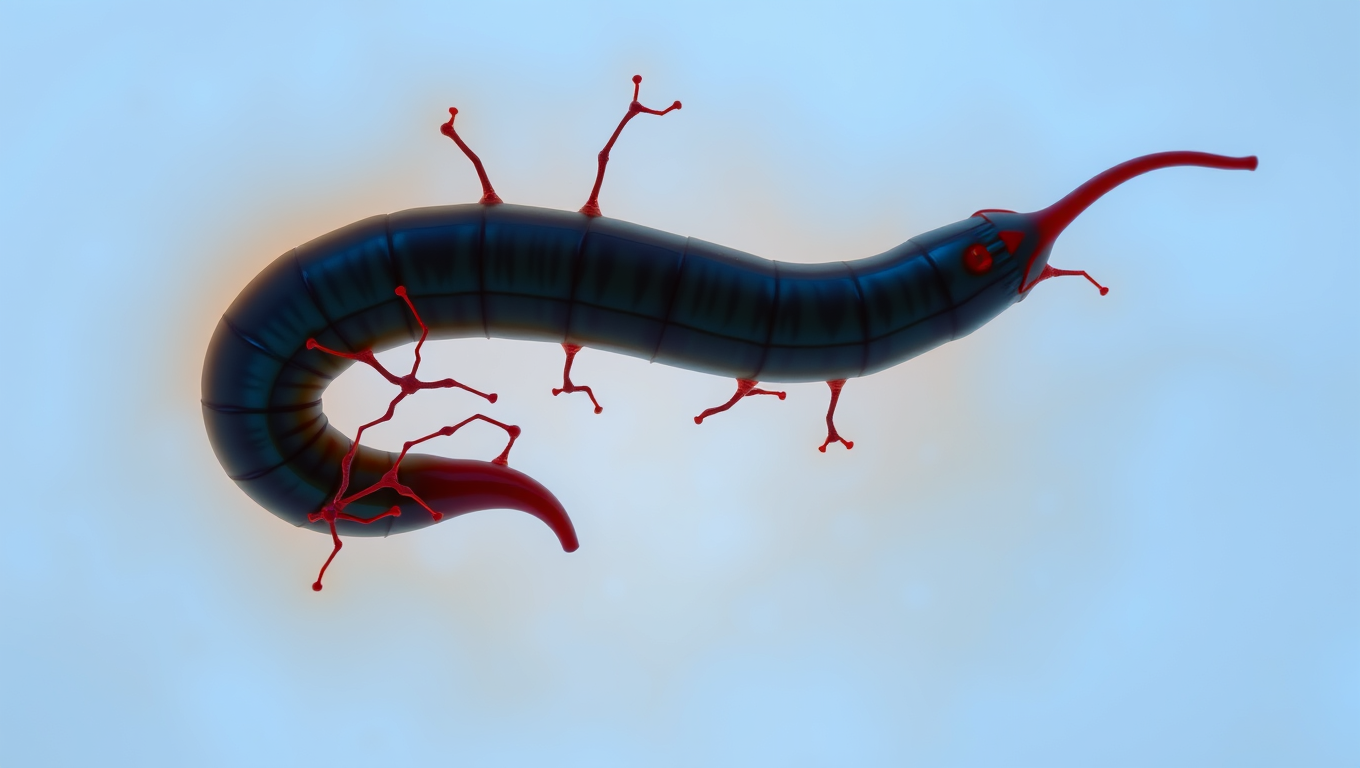
“The Silent Invader: How a Parasitic Worm Evades Detection and What it Can Teach Us About Pain Relief”
Scientists have discovered a parasite that can sneak into your skin without you feeling a thing. The worm, Schistosoma mansoni, has evolved a way to switch off the body’s pain and itch signals, letting it invade undetected. By blocking certain nerve pathways, it avoids triggering the immune system’s alarms. This stealth tactic not only helps the worm survive, but could inspire new kinds of pain treatments and even preventative creams to protect people from infection.
Bacteria
Unlocking the Secrets of Mars: Cosmic Rays Reveal Hidden Potential for Life
Cosmic rays from deep space might be the secret energy source that allows life to exist underground on Mars and icy moons like Enceladus and Europa. New research reveals that when these rays interact with water or ice below the surface, they release energy-carrying electrons that could feed microscopic life, a process known as radiolysis. This breakthrough suggests that life doesn’t need sunlight or heat, just some buried water and radiation.
Alternative Medicine
Cleaning Up the Water, Cooling Down the Risks: A New Approach to Safer Romaine Lettuce
Romaine lettuce has a long history of E. coli outbreaks, but scientists are zeroing in on why. A new study reveals that the way lettuce is irrigated—and how it’s kept cool afterward—can make all the difference. Spraying leaves with untreated surface water is a major risk factor, while switching to drip or furrow irrigation cuts contamination dramatically. Add in better cold storage from harvest to delivery, and the odds of an outbreak plummet. The research offers a clear, science-backed path to safer salads—one that combines smarter farming with better logistics.
-

 Detectors10 months ago
Detectors10 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate11 months ago
Earth & Climate11 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer10 months ago
Cancer10 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein11 months ago
Albert Einstein11 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry10 months ago
Chemistry10 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate10 months ago
Earth & Climate10 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food10 months ago
Agriculture and Food10 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions11 months ago
Diseases and Conditions11 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention