While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Environmental Policies
A Flawed Model Threatens COVID-19 Research Progress
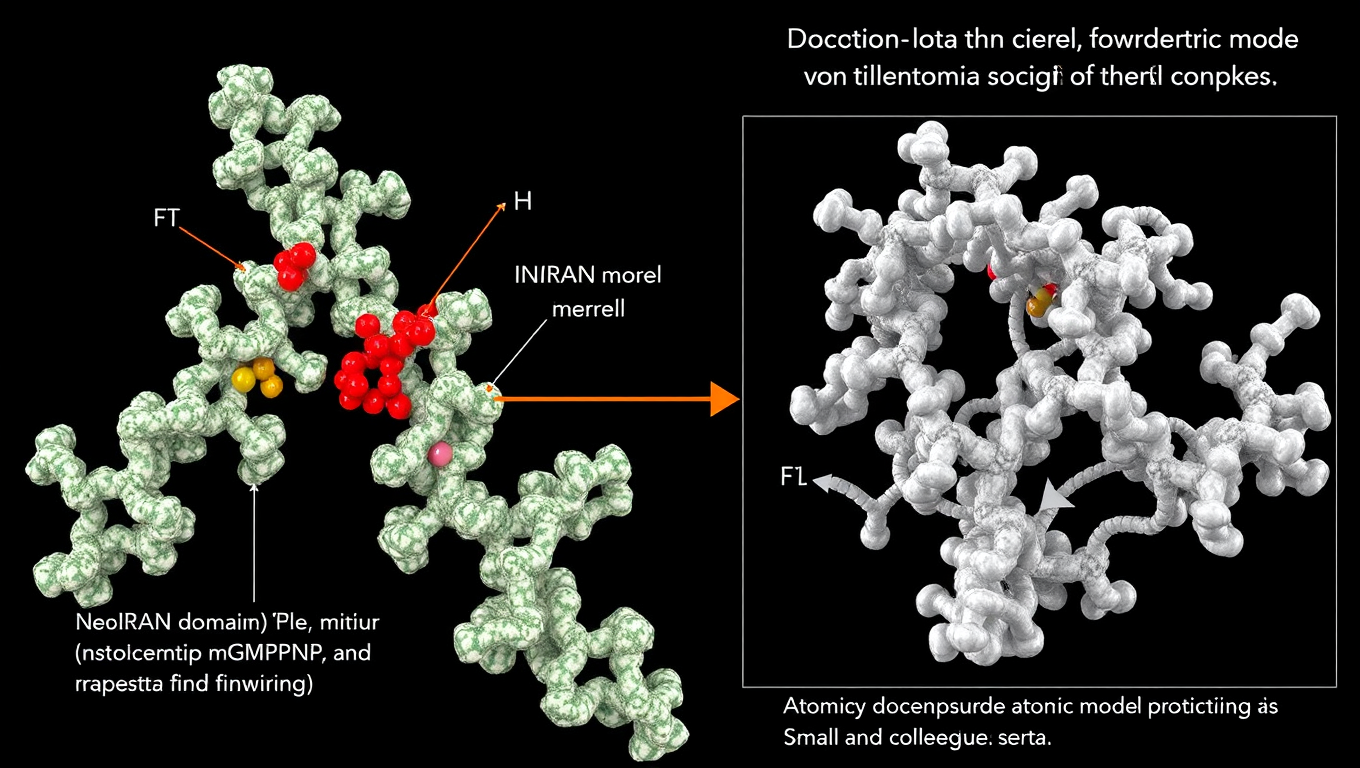
A promising path to fighting COVID and other coronaviruses may have been based on a serious mistake. Scientists had zeroed in on a part of the virus called the NiRAN domain, believed to be a powerful target for new antiviral drugs. But when a Rockefeller team revisited a highly cited 2022 study, they found the evidence didn’t hold up. Key molecules shown in the original virus model were actually missing. Their discovery could help prevent wasted time and resources in the race to develop better treatments—and highlights how even one bad blueprint can throw off years of research.
Educational Policy
700,000 Years Ahead of Their Teeth: The Carbs That Made Us Human
Long before evolution equipped them with the right teeth, early humans began eating tough grasses and starchy underground plants—foods rich in energy but hard to chew. A new study reveals that this bold dietary shift happened 700,000 years before the ideal dental traits evolved to handle it.
Cancer
Safer Non-Stick Coatings: Scientists Develop Alternative to Teflon
Scientists at the University of Toronto have developed a new non-stick material that rivals the performance of traditional PFAS-based coatings while using only minimal amounts of these controversial “forever chemicals.” Through an inventive process called “nanoscale fletching,” they modified silicone-based polymers to repel both water and oil effectively. This breakthrough could pave the way for safer cookware, fabrics, and other products without the environmental and health risks linked to long-chain PFAS.
Depression
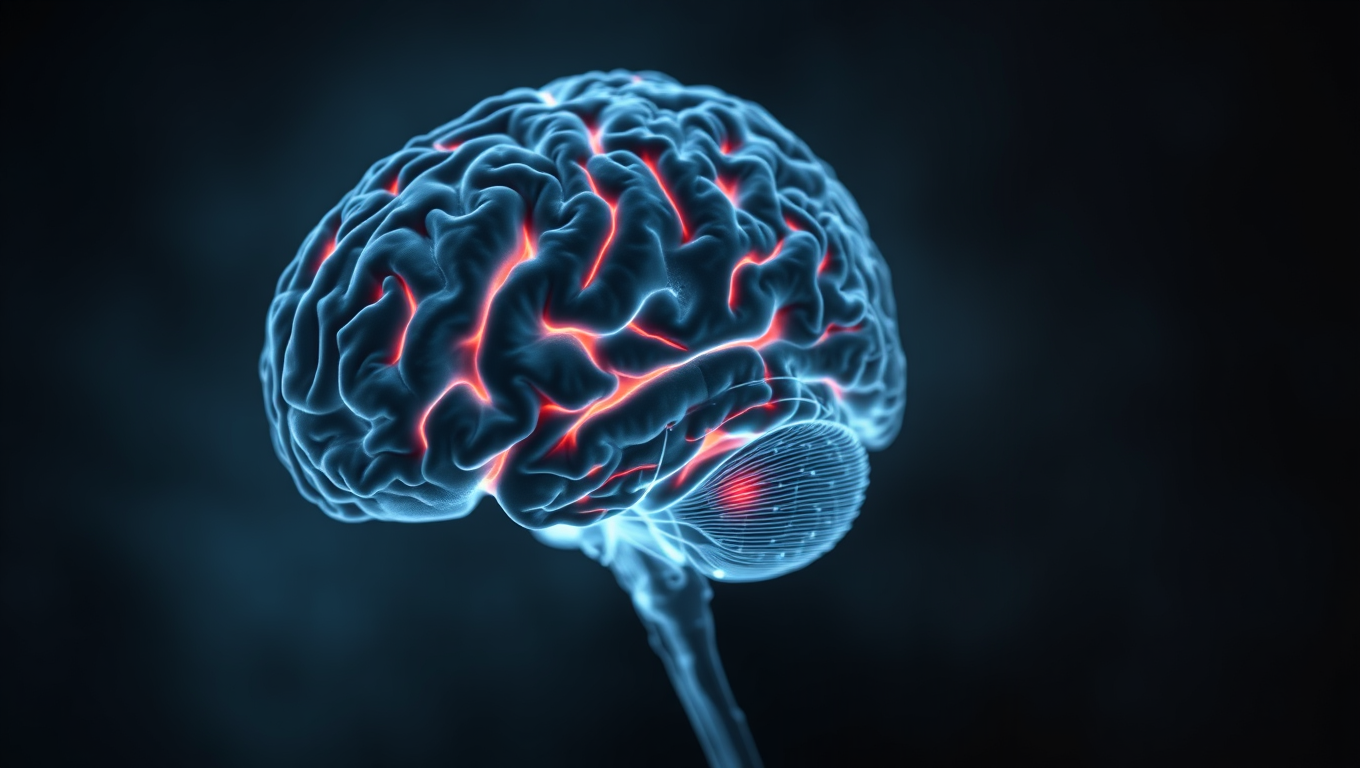
The Unseen Toll of the Pandemic: How Stress and Isolation May Be Aging Your Brain
Even people who never caught Covid-19 may have aged mentally faster during the pandemic, according to new brain scan research. This large UK study shows how the stress, isolation, and upheaval of lockdowns may have aged our brains, especially in older adults, men, and disadvantaged individuals. While infection itself impacted some thinking skills, even those who stayed virus-free showed signs of accelerated brain aging—possibly reversible. The study highlights how major life disruptions, not just illness, can reshape our mental health.
-

 Detectors10 months ago
Detectors10 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate12 months ago
Earth & Climate12 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer11 months ago
Cancer11 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein12 months ago
Albert Einstein12 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry11 months ago
Chemistry11 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate11 months ago
Earth & Climate11 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food11 months ago
Agriculture and Food11 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions12 months ago
Diseases and Conditions12 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention