While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Bird Flu Research
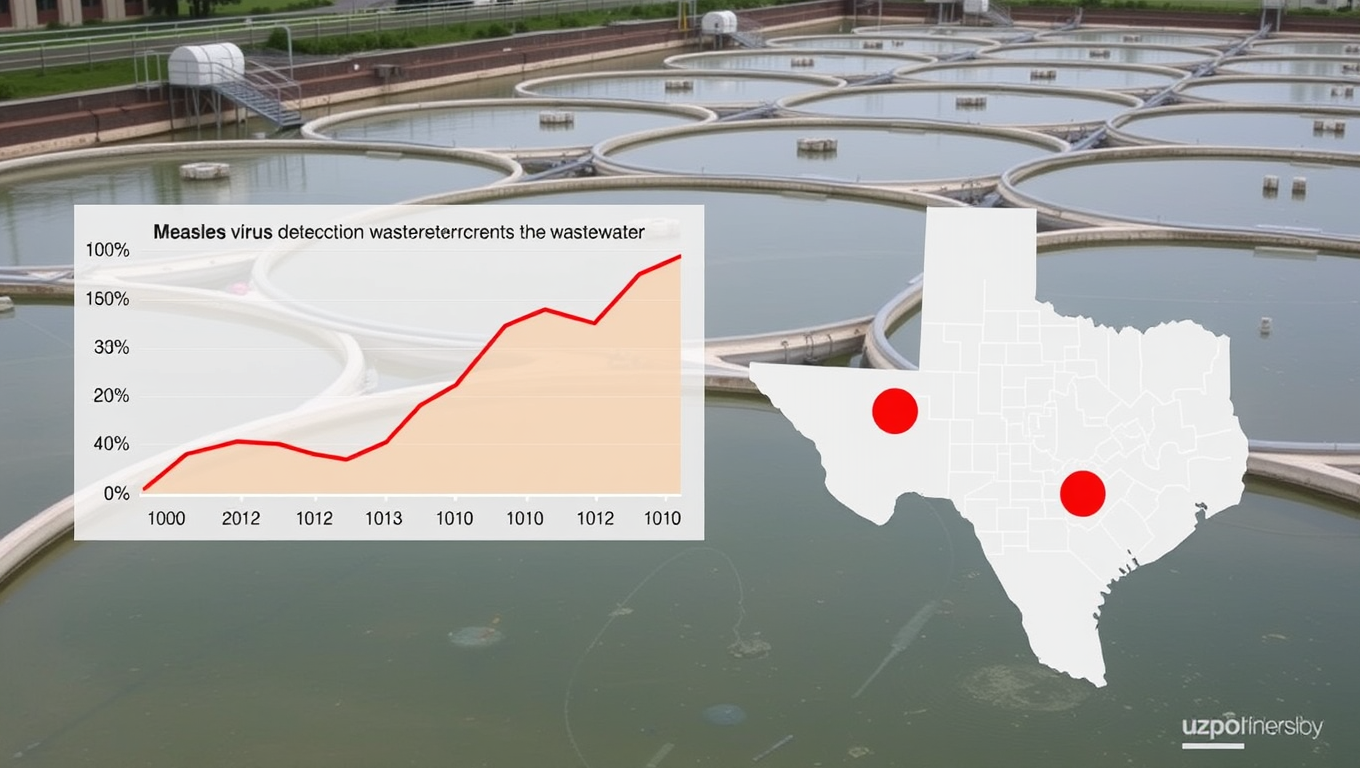
Measles Virus Detected in Houston Wastewater Before Cases Were Reported: A Promising Public Health Strategy
An innovative outbreak detection program that tracks disease-causing viruses in wastewater identified the measles virus in Houston samples collected in early January 2025, before cases were reported.
Bird Flu Research
HIV Discovery Could Open Door to Long-Sought Cure
New HIV research shows that small changes in the virus affect how quickly or slowly it replicates and how easily it can reawaken in the body. These insights bring researchers closer to finding ways to flush out the dormant virus and eliminate it for good.
Bird Flu
The Evolution of a Single Gene Allowed the Plague to Adapt and Survive for Centuries
Scientists have documented the way a single gene in the bacterium that causes bubonic plague, Yersinia pestis, allowed it to survive hundreds of years by adjusting its virulence and the length of time it took to kill its victims, but these forms of plague ultimately died out.
Bird Flu Research
“Ancient Arctic Nursery: 73 Million-Year-Old Bird Fossils Discovered in Alaska”
Spring in the Arctic brings forth a plethora of peeps and downy hatchlings as millions of birds gather to raise their young. The same was true 73 million years ago, according to a new article. The paper documents the earliest-known example of birds nesting in the polar regions.
-

 Detectors11 months ago
Detectors11 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate12 months ago
Earth & Climate12 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer12 months ago
Cancer12 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein12 months ago
Albert Einstein12 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry11 months ago
Chemistry11 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate12 months ago
Earth & Climate12 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Diseases and Conditions12 months ago
Diseases and Conditions12 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention
-

 Agriculture and Food12 months ago
Agriculture and Food12 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”