While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Early Humans
Revolutionizing Species Tree Inference with ROADIES
Engineers are making it easier for researchers from a broad range of backgrounds to understand how different species are evolutionarily related, and support the transformative biological and medical applications that rely on these species trees. The researchers developed a scalable, automated and user-friendly tool called ROADIES that allows scientists to infer species trees directly from raw genome data, with less reliance on the domain expertise and computational resources currently required.
Early Humans
The Hidden Legacy of the Denisovans: Uncovering the Secrets of Human Evolution
Denisovans, a mysterious human relative, left behind far more than a handful of fossils—they left genetic fingerprints in modern humans across the globe. Multiple interbreeding events with distinct Denisovan populations helped shape traits like high-altitude survival in Tibetans, cold-weather adaptation in Inuits, and enhanced immunity. Their influence spanned from Siberia to South America, and scientists are now uncovering how these genetic gifts transformed human evolution, even with such limited physical remains.
Early Humans
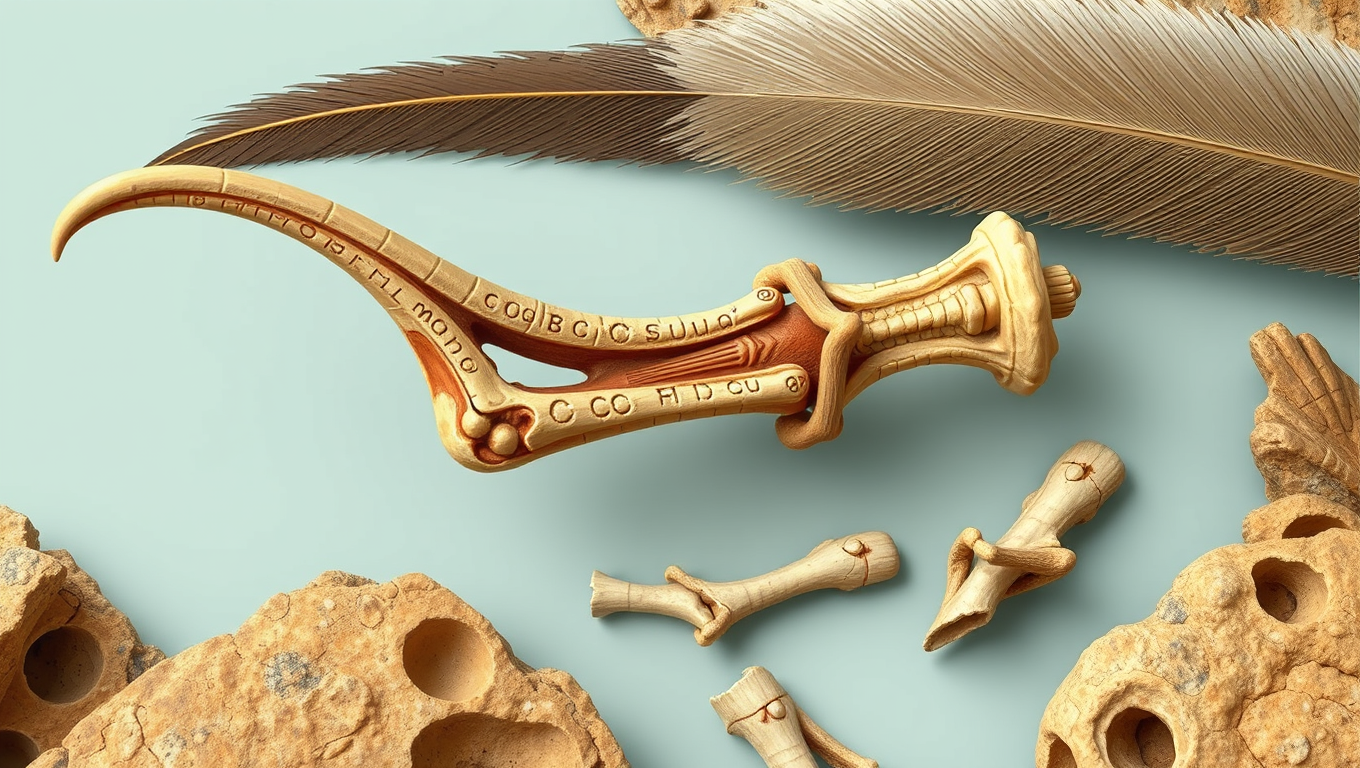
A Tiny Dinosaur Bone Rewrites the Origin of Bird Flight
A tiny, overlooked wrist bone called the pisiform may have played a pivotal role in bird flight and it turns out it evolved far earlier than scientists thought. Fossils from bird-like dinosaurs in Mongolia reveal that this bone, once thought to vanish and reappear, was actually hiding in plain sight. Thanks to pristine preservation and 3D scans, researchers connected the dots between ancient theropods and modern birds, uncovering a deeper, more intricate story of how dinosaurs evolved the tools for powered flight.
Ancient Civilizations
Unraveling a 130-Year-Old Literary Mystery: The Song of Wade Finally Solved
After baffling scholars for over a century, Cambridge researchers have reinterpreted the long-lost Song of Wade, revealing it to be a chivalric romance rather than a monster-filled myth. The twist came when “elves” in a medieval sermon were correctly identified as “wolves,” dramatically altering the legend’s tone and context.
-

 Detectors11 months ago
Detectors11 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate12 months ago
Earth & Climate12 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer12 months ago
Cancer12 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein12 months ago
Albert Einstein12 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry11 months ago
Chemistry11 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate11 months ago
Earth & Climate11 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food12 months ago
Agriculture and Food12 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions12 months ago
Diseases and Conditions12 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention