While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Ebola
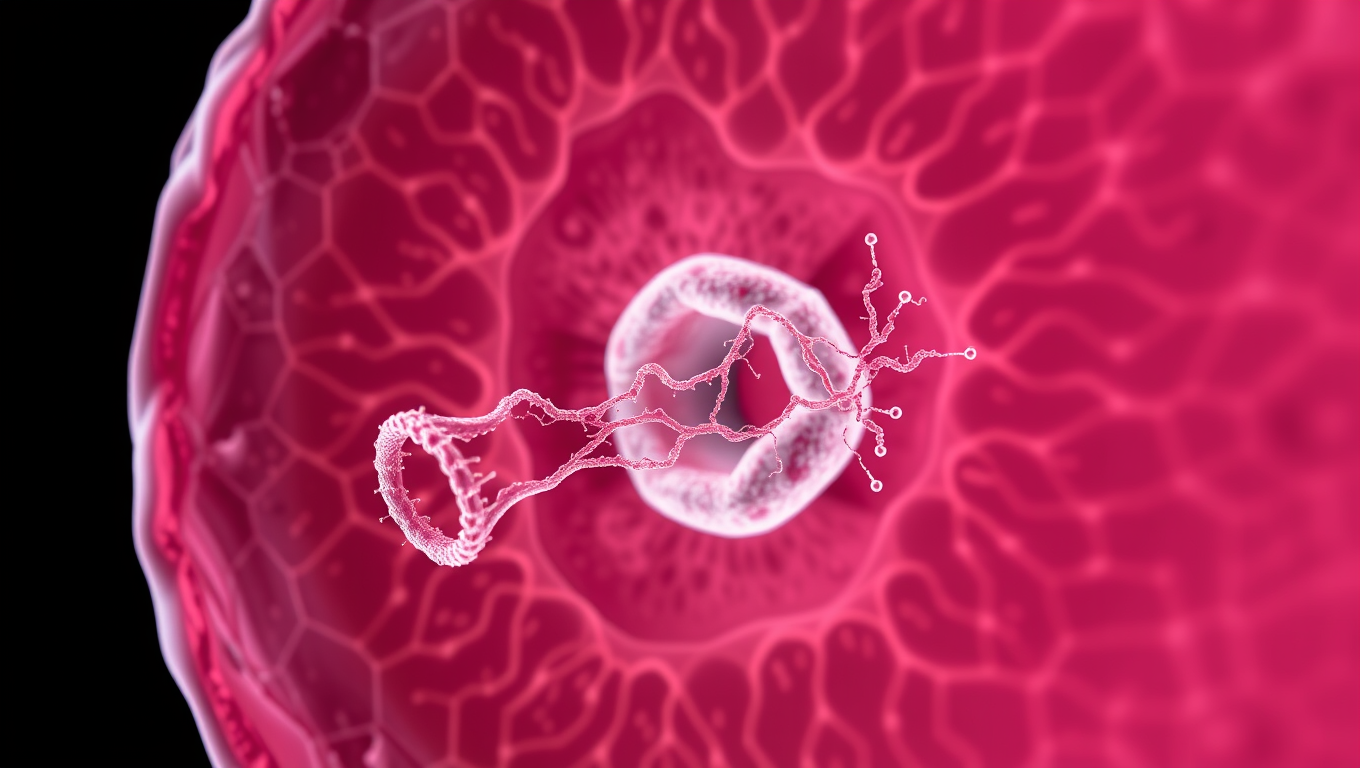
Unlocking Zika’s Secrets: How a Virus Builds Tunnels to Infect Fetal Cells
The Zika virus builds tiny tunnels, called tunneling nanotubes, to stealthily transport material needed to infect nearby cells, including in placental cells, according to a team of researchers from Penn State and Baylor College of Medicine. It’s one way the virus crosses the placental barrier, transmitting from mother to fetus during pregnancy without raising alarm in the immune system. The team also demonstrated, for the first time, that one specific Zika protein — non-structural protein 1 (NS1) — is responsible for the formation of the nanotubes.
Cold and Flu
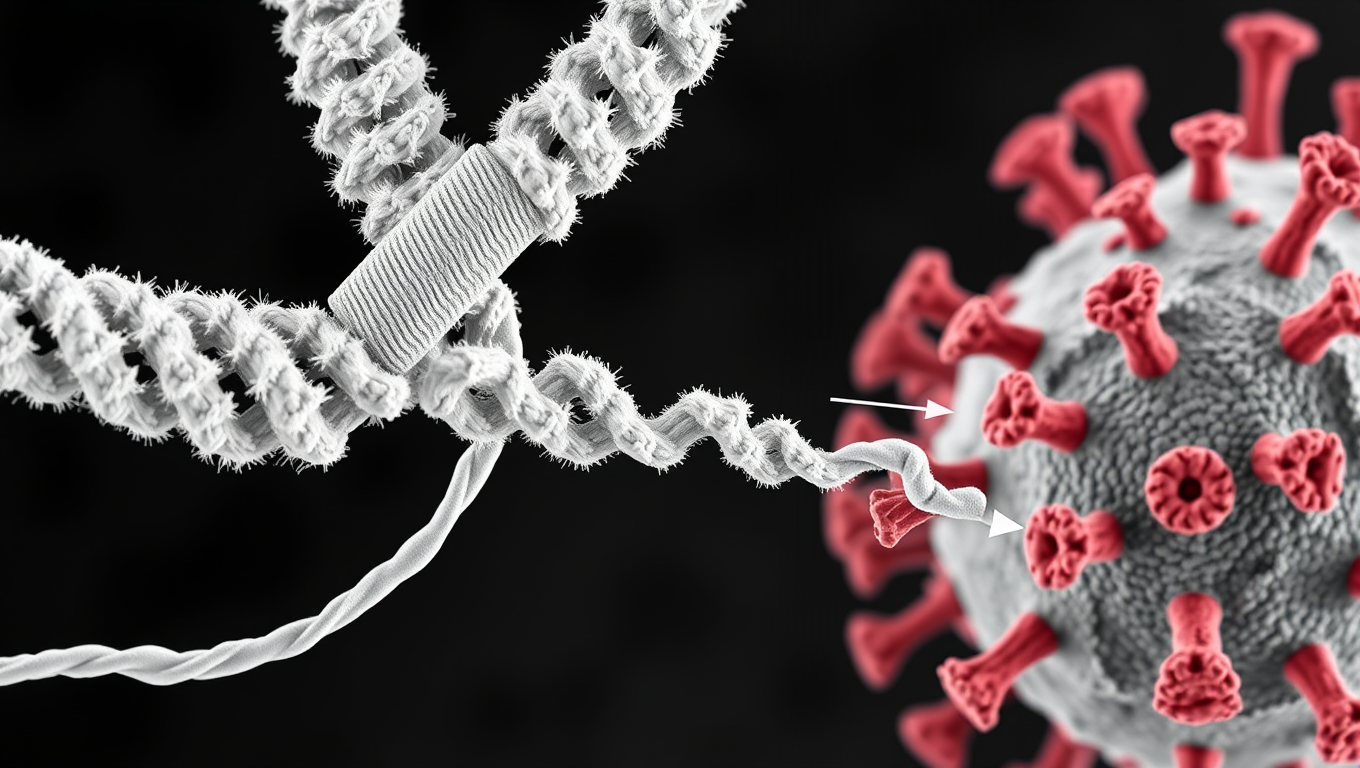
Scientists Discover Llama Antibodies That Shut Down COVID-19 and Its Future Variants
Powerful llama-derived antibodies could be the key to stopping not just current SARS viruses, but future ones too. Scientists have discovered a unique class of nanobodies that clamp the coronavirus spike protein shut at a highly conserved region, rendering it unable to infect cells. Unlike existing therapies that target mutating regions, this approach strikes at the virus s core machinery, giving it little room to evolve. Even when pushed to mutate, the virus faltered, making this a high-potential strategy for broad, lasting protection.
Bird Flu Research
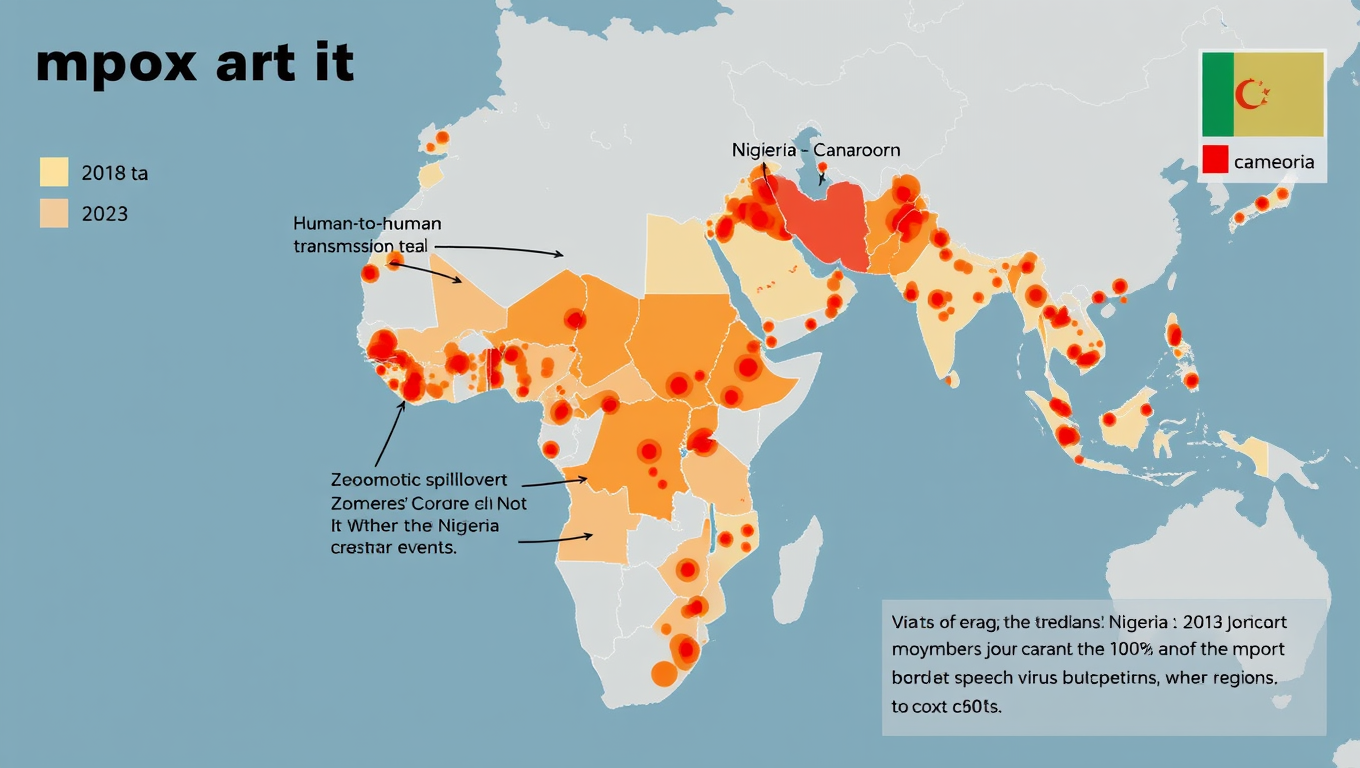
Widespread Mpox Transmission in West Africa Before 2022 Outbreak Revealed by Genomic Data
Historically, most human mpox infections have resulted from zoonotic transmission –m eaning from animals to humans — and these spillovers have rarely led to human-to-human transmission. But during the 2022 global outbreak, mpox began spreading readily between people. A new study now shows the virus was circulating long before then.
Ebola
“Unveiling the Hidden Threat: A Deeper Dive into Hantavirus and Its Rodent Hosts”
Virginia Tech researchers seek to understand the environmental factors that influence the distribution of hantavirus in rodent populations across the United States.
-

 Detectors10 months ago
Detectors10 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate11 months ago
Earth & Climate11 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer10 months ago
Cancer10 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein11 months ago
Albert Einstein11 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry10 months ago
Chemistry10 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate10 months ago
Earth & Climate10 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food11 months ago
Agriculture and Food11 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions11 months ago
Diseases and Conditions11 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention