While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Biology
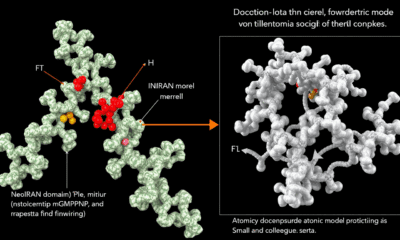
Unlocking the Secrets of Protein Synthesis: A New Toolbox for Characterizing Internal Ribosomal Entry Sites (IRESes)
The research field of ‘cellular IRESes’ lay dormant for decades, as there was no uniform standard of reliable methods for the clear characterization of these starting points for the ribosome-mediated control of gene expression. Researchers have now developed a toolbox as a new gold standard for this field. They hope to discover strong IRES elements that are directly relevant for synthetic biology and for application in emerging mRNA therapeutics.

Biochemistry Research
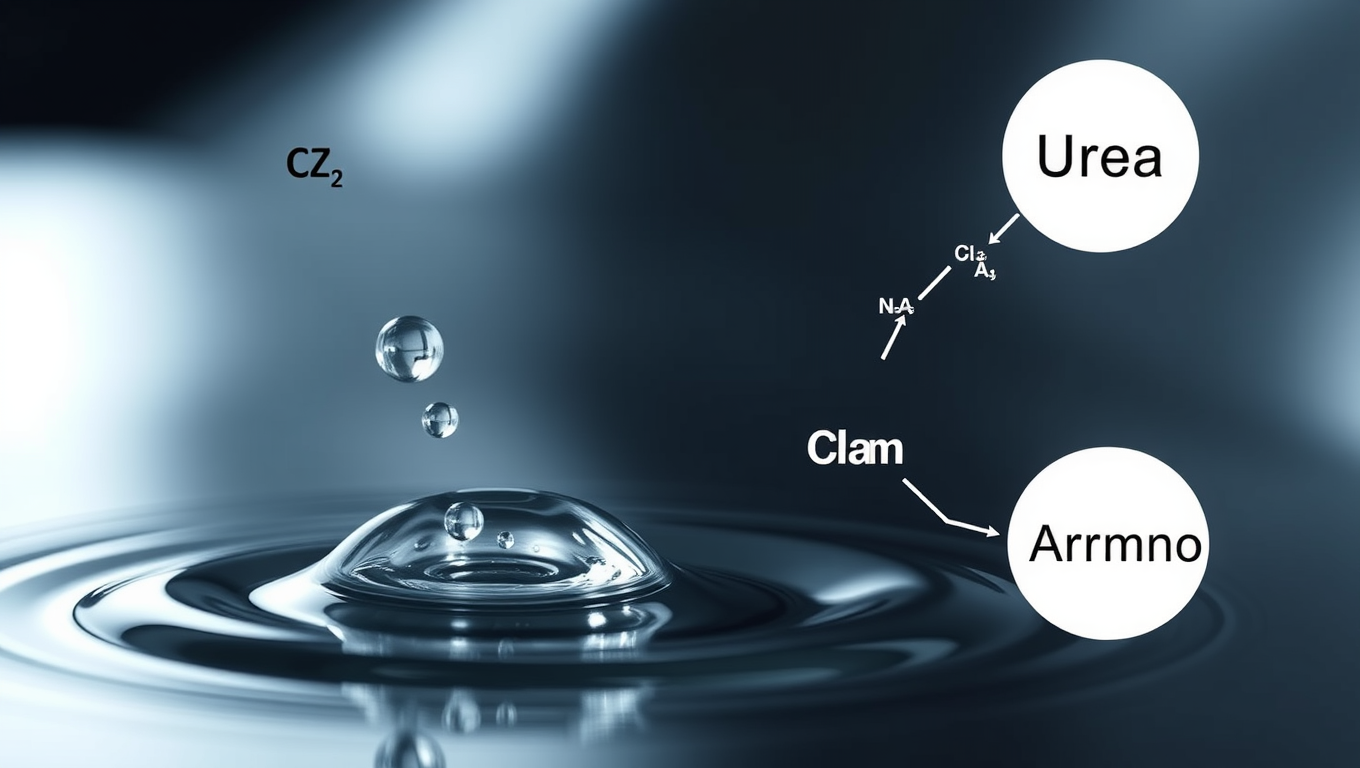
Unlocking the Secrets of Life: A Spontaneous Reaction that Could Have Started it All
Scientists have uncovered a surprising new way that urea—an essential building block for life—could have formed on the early Earth. Instead of requiring high temperatures or complex catalysts, this process occurs naturally on the surface of tiny water droplets like those in sea spray or fog. At this boundary between air and water, a unique chemical environment allows carbon dioxide and ammonia to combine and spontaneously produce urea, without any added energy. The finding offers a compelling clue in the mystery of life’s origins and hints that nature may have used simple, everyday phenomena to spark complex biological chemistry.
Bacteria
Unveiling the Secrets of Pandoraea: How Lung Bacteria Forge Iron-Stealing Weapons to Survive
Researchers investigating the enigmatic and antibiotic-resistant Pandoraea bacteria have uncovered a surprising twist: these pathogens don’t just pose risks they also produce powerful natural compounds. By studying a newly discovered gene cluster called pan, scientists identified two novel molecules Pandorabactin A and B that allow the bacteria to steal iron from their environment, giving them a survival edge in iron-poor places like the human body. These molecules also sabotage rival bacteria by starving them of iron, potentially reshaping microbial communities in diseases like cystic fibrosis.
Animals
“Red Vision Unlocked: Mediterranean Beetles Shatter Insect Color Limitations”
Beetles that can see the color red? That s exactly what scientists discovered in two Mediterranean species that defy the norm of insect vision. While most insects are blind to red, these beetles use specialized photoreceptors to detect it and even show a strong preference for red flowers like poppies and anemones. This breakthrough challenges long-standing assumptions about how flower colors evolved and opens a new path for studying how pollinators influence plant traits over time.
-

 Detectors3 months ago
Detectors3 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate4 months ago
Earth & Climate4 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer4 months ago
Cancer4 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Agriculture and Food4 months ago
Agriculture and Food4 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions4 months ago
Diseases and Conditions4 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention
-

 Chemistry3 months ago
Chemistry3 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Albert Einstein4 months ago
Albert Einstein4 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Earth & Climate3 months ago
Earth & Climate3 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals