While we try to keep things accurate, this content is part of an ongoing experiment and may not always be reliable.
Please double-check important details — we’re not responsible for how the information is used.
Charles Darwin
Unveiling the Secrets of Ice Age Evolution: A Study of Adaptation and Survival
Cold-adapted animals started to evolve 2.6 million years ago when the permanent ice at the poles became more prevalent. There followed a time when the continental ice sheets expanded and contracted and around 700,000 years ago the cold periods doubled in length. This is when many of the current cold-adapted species, as well as extinct ones like mammoths, evolved.

Animals
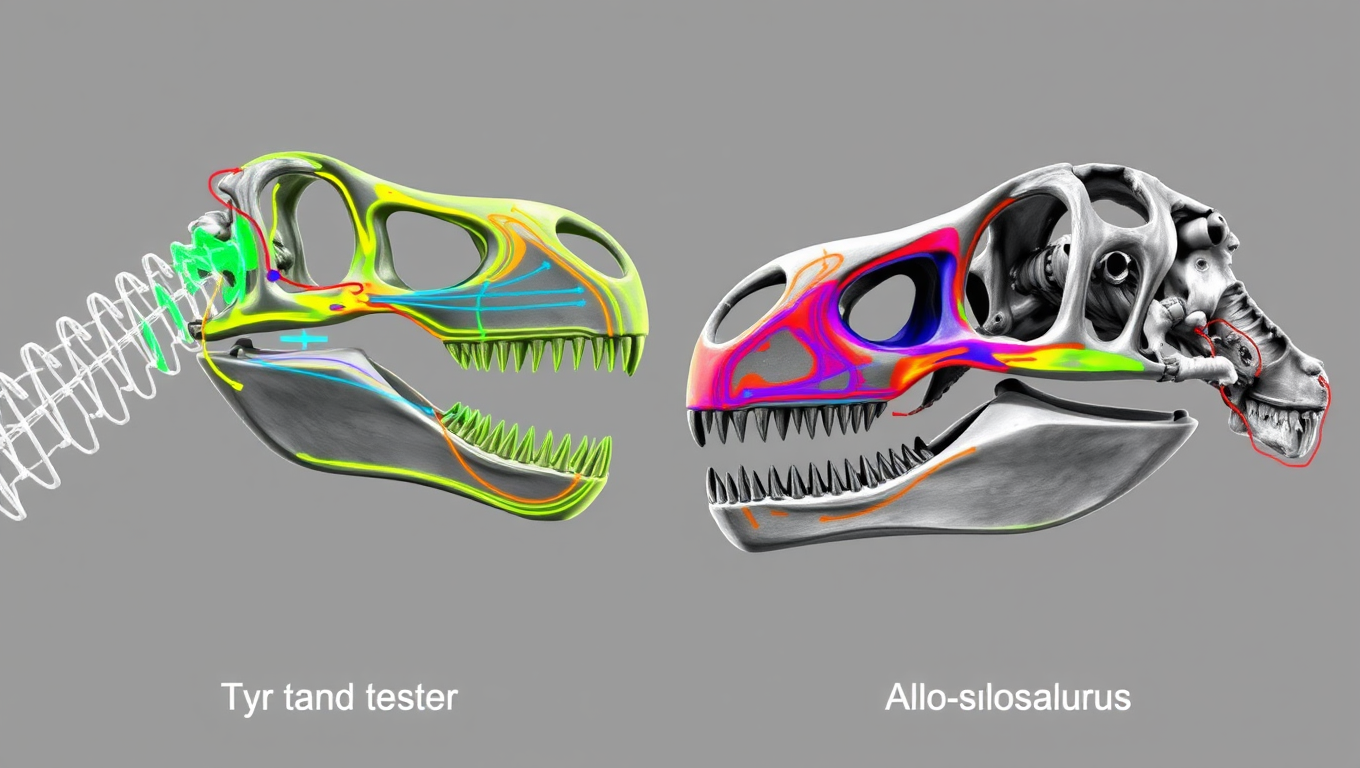
Crushing vs. Slashing: New Skull Scans Reveal How Giant Dinosaurs Hunted Prey
Tyrannosaurus rex might be the most famous meat-eater of all time, but it turns out it wasn’t the only way to be a terrifying giant. New research shows that while T. rex evolved a skull designed for bone-crushing bites like a modern crocodile, other massive carnivorous dinosaurs like spinosaurs and allosaurs took a very different route — specializing in slashing and tearing flesh instead.
Charles Darwin
The 10,000-mile March Through Fire: How Dinosaurs Evolved to Thrive
Despite Earth’s most devastating mass extinction wiping out over 80% of marine life and half of land species, a group of early reptiles called archosauromorphs not only survived but thrived, venturing across the supposedly lifeless tropics to eventually evolve into the dinosaurs and crocodiles we know today. Armed with a groundbreaking model dubbed TARDIS, researchers have reconstructed their ancient dispersal routes, revealing how these resilient reptiles conquered a hostile, post-apocalyptic Earth.
Ancient DNA
Chemists Recreate a Crucial Step in the Origin of Life
Chemists have demonstrated how RNA (ribonucleic acid) might have replicated itself on early Earth — a key process in the origin of life.
-

 Detectors11 months ago
Detectors11 months agoA New Horizon for Vision: How Gold Nanoparticles May Restore People’s Sight
-

 Earth & Climate12 months ago
Earth & Climate12 months agoRetiring Abroad Can Be Lonely Business
-

 Cancer12 months ago
Cancer12 months agoRevolutionizing Quantum Communication: Direct Connections Between Multiple Processors
-

 Albert Einstein12 months ago
Albert Einstein12 months agoHarnessing Water Waves: A Breakthrough in Controlling Floating Objects
-

 Chemistry12 months ago
Chemistry12 months ago“Unveiling Hidden Patterns: A New Twist on Interference Phenomena”
-

 Earth & Climate12 months ago
Earth & Climate12 months agoHousehold Electricity Three Times More Expensive Than Upcoming ‘Eco-Friendly’ Aviation E-Fuels, Study Reveals
-

 Agriculture and Food12 months ago
Agriculture and Food12 months ago“A Sustainable Solution: Researchers Create Hybrid Cheese with 25% Pea Protein”
-

 Diseases and Conditions12 months ago
Diseases and Conditions12 months agoReducing Falls Among Elderly Women with Polypharmacy through Exercise Intervention